When you take multiple medications, the risk of harmful interactions goes up-not just because of the drugs themselves, but because of your genes. Two people taking the exact same pills at the same doses can have wildly different outcomes. One feels fine. The other ends up in the hospital. Why? It’s not random. It’s pharmacogenomics.
What Pharmacogenomics Actually Means
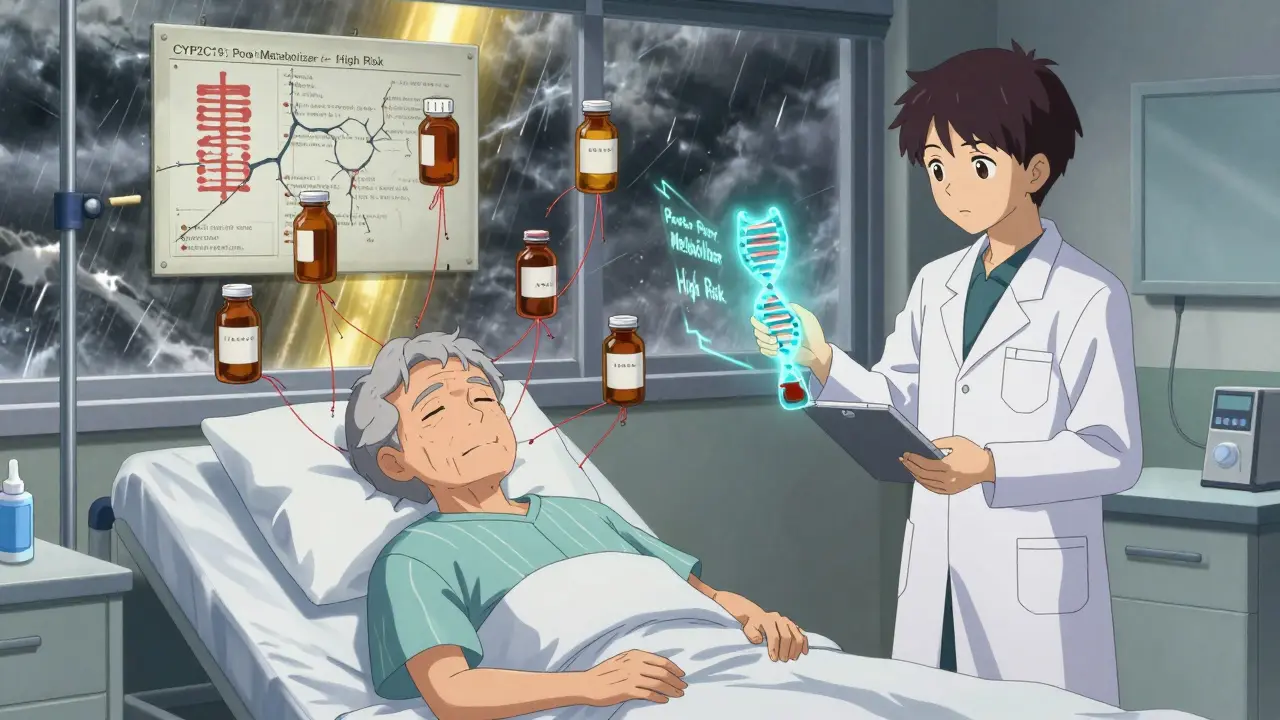
Pharmacogenomics is the study of how your genes affect how your body responds to drugs. It’s not about whether you’re allergic to penicillin. It’s about whether your liver can break down a drug fast enough, or too slowly, because of the version of a gene you inherited. This isn’t science fiction. It’s in the FDA’s official guidelines. Over 300 drugs now include pharmacogenomic information on their labels. That includes common ones like warfarin, clopidogrel, statins, antidepressants, and painkillers.For example, if you’re a poor metabolizer of CYP2D6-a gene that helps break down about 25% of all prescription drugs-you might get dangerously high levels of codeine or tramadol even at normal doses. Your body can’t convert them properly, so they build up. That’s not a drug interaction. That’s a gene-drug interaction. And it’s completely invisible to standard drug interaction checkers.
How Your Genes Change Drug Interaction Risk
Most drug interaction tools only look at two drugs. They say, “This antibiotic and this blood thinner shouldn’t be taken together.” But they don’t know if you’re a fast, normal, or slow metabolizer. That’s like checking for traffic lights but ignoring whether your car’s brakes work.There are three main ways genes change how drugs interact:
- Inhibitory interactions: One drug blocks the enzyme that breaks down another. If you’re already a slow metabolizer because of your genes, this can push drug levels into toxic range.
- Induction interactions: One drug speeds up enzyme activity. If you’re a fast metabolizer, this might make your medication stop working entirely.
- Phenoconversion: A drug temporarily turns your body into a different metabolic type. A normal metabolizer could act like a poor metabolizer just because they’re taking another medication.
Take antidepressants and beta-blockers together. On paper, it’s a low-risk combo. But if you’re a CYP2D6 poor metabolizer, and you’re also taking fluoxetine (which blocks CYP2D6), you’re getting a double hit. Your body can’t clear either drug. Blood levels spike. Side effects like dizziness, heart rhythm changes, or even seizures become real risks.
Why Traditional Drug Checkers Miss the Mark
Standard databases like Lexicomp or Micromedex list around 50,000 potential drug interactions. But they ignore genetics. A 2022 study in the American Journal of Managed Care found that when you add genetic data, the number of clinically relevant interactions jumps by 34%. In some cases, the estimated risk of major interactions went up by over 90%.That’s not a small correction. It’s a complete overhaul of how we think about safety. A patient on five medications might be flagged for two interactions by a standard checker. Add their CYP2C19 and CYP2D6 status, and suddenly they’re at risk for five major interactions-three of which were completely invisible before.
Antidepressants, antipsychotics, and pain meds are the biggest culprits. Why? Because they’re often metabolized by just one or two enzymes-and those enzymes are the same ones affected by other common drugs. CYP2D6 and CYP2C19 are the most common troublemakers. If you’re on a statin, an SSRI, and a beta-blocker, and you’re a slow metabolizer for both enzymes, you’re walking a tightrope.
Real-World Impact: Numbers That Matter
Adverse drug reactions cost the U.S. healthcare system $30 billion a year. One in five hospitalizations in older adults is caused by a drug reaction. And nearly half of those could be prevented if we knew the patient’s genetic profile.At Mayo Clinic, where they’ve been testing patients preemptively since 2011, 89% of people had at least one actionable gene variant. When they added genetic alerts to their electronic health records, inappropriate prescribing dropped by 45%. That’s not theory. That’s real patients avoiding falls, kidney damage, and heart problems.
For warfarin, the classic example, PGx-guided dosing cuts major bleeding events by 31%. For clopidogrel, patients with a CYP2C19 loss-of-function variant get no benefit from the drug at all-unless you switch them to prasugrel or ticagrelor. That’s life-saving.
Where It’s Working-and Where It’s Not
Some places are ahead. Vanderbilt’s PREDICT program has tested over 100,000 patients. Mayo Clinic, Stanford, and Cleveland Clinic all have PGx programs built into routine care. But outside academic centers? It’s still rare.Only 15% of U.S. healthcare systems have PGx results integrated into their electronic records. Only 28% of pharmacists feel trained to interpret the results. And even when tests are done, doctors often don’t know what to do with them. A 2023 survey found 67% of pharmacists said lack of clinical decision support was a major barrier.
The guidelines exist. The Clinical Pharmacogenetics Implementation Consortium (CPIC) has clear, evidence-based rules for over 100 gene-drug pairs. But they’re not widely used. Why? Because it’s not just about the science. It’s about systems. It takes time, training, and money-about $1.2 million per hospital to fully implement, according to one study.
The Big Gaps: Diversity and Access
Here’s the uncomfortable truth: most of the data comes from people of European ancestry. Only 2% of pharmacogenomics research participants are of African descent. That means the guidelines we use might not work for everyone.For example, the HLA-B*15:02 variant, which puts people at extreme risk of deadly skin reactions from carbamazepine, is common in Southeast Asian populations but rare in Europeans. If you only test based on European data, you miss the risk. That’s not just a gap. It’s a danger.
Reimbursement is another wall. Only 19 CPT codes exist for PGx testing. Insurance often won’t pay unless it’s a “companion diagnostic”-meaning it’s tied to a specific drug, like testing for EGFR mutations before giving lung cancer drugs. For routine use? You’re often paying out of pocket. Tests cost $250-$400. Not cheap, but far cheaper than an ER visit for a drug reaction.
What’s Next
The FDA plans to add 24 new gene-drug pairs to its list in 2024. The NIH’s All of Us program has already returned PGx results to over 250,000 people. AI models are starting to combine genetic data with drug lists to predict interactions with 37% more accuracy than old methods.The goal isn’t to test everyone tomorrow. It’s to make it routine for people on multiple medications, especially those over 65, with chronic conditions, or in psychiatric care. That’s where the risk is highest.
Pharmacogenomics doesn’t eliminate drug interactions. It makes them predictable. And that’s the difference between guessing and knowing. Between harm and safety. Between standard care and personalized care.
If you’re on five or more drugs, ask your doctor: “Has my genetic profile been considered?” If they don’t know what you mean, ask for a referral to a clinical pharmacist who specializes in pharmacogenomics. You’re not asking for a luxury. You’re asking for a basic safety check.





Comments (12)
McCarthy Halverson
January 11, 2026 AT 19:07 PMBeen on 6 meds for years. Got tested last year. Turned out I’m a CYP2D6 poor metabolizer. My doc switched my tramadol to gabapentin. No more dizziness. Simple fix, if you know to ask.
Michael Marchio
January 11, 2026 AT 22:31 PMLet me break this down for you because clearly no one else is willing to. The entire system is broken. We’re treating people like lab rats while pharmaceutical companies profit off the ignorance of clinicians. The FDA lists 300+ drugs with PGx info but 90% of prescribers don’t know what CYP2D6 even is. It’s not negligence-it’s institutionalized incompetence. And don’t get me started on how insurance refuses to cover testing unless you’re dying. This isn’t medicine. It’s a lottery where your genes decide if you live or end up in the ER.
Jake Kelly
January 13, 2026 AT 16:30 PMReally appreciate this post. I work in geriatrics and see this every week. Grandpa on 8 meds, no genetic data, ends up in the hospital for something that could’ve been avoided. We need more pharmacists involved early. Not just after the fact.
Ashlee Montgomery
January 15, 2026 AT 00:27 AMIt’s funny how we’ve spent decades trying to perfect drug dosing based on weight, age, kidney function-but never asked what’s inside the person’s DNA. We treat the body like a machine with universal parts. But it’s not. It’s a unique biological signature. Why did it take so long to realize that?
neeraj maor
January 16, 2026 AT 02:44 AMThey’re hiding something. PGx testing is expensive. Who benefits? Labs. Pharma. Big Med. Why are they pushing this now? Because they can charge more. And the ‘clinical evidence’? All funded by the same companies selling the tests. I’ve seen patients get flagged for ‘risk’ then get prescribed the same drug anyway. This is a money scheme wrapped in science.
Ritwik Bose
January 17, 2026 AT 04:55 AMThank you for sharing this deeply important perspective 🙏
As someone from India, I’ve seen how access to such knowledge is limited, yet the need is immense. We must advocate for equitable implementation-not just in the US but globally. PGx isn’t a privilege. It’s a right. Let’s ensure no one is left behind because of geography or cost.
Paul Bear
January 19, 2026 AT 03:02 AMThere’s a fundamental epistemological flaw in the current paradigm: pharmacokinetic variability is treated as noise rather than signal. The CYP450 isoforms-particularly CYP2D6 and CYP2C19-are polymorphic in a clinically significant manner, yet most EHRs lack the structural integration to interpret diplotype data. This isn’t just an oversight-it’s a systemic failure of translational informatics. The CPIC guidelines are robust, but without automated clinical decision support, they’re just PDFs on a server.
Jaqueline santos bau
January 20, 2026 AT 23:17 PMMy cousin took fluoxetine and metoprolol together and ended up in ICU with torsades de pointes. They didn’t test her genes. She was 32. Now she’s on disability. This isn’t ‘science.’ It’s negligence. And no one’s held accountable. Why? Because doctors don’t want to admit they don’t know this stuff.
Kunal Majumder
January 20, 2026 AT 23:41 PMBro, I got tested after my dad had a bad reaction. Turned out I’m a slow metabolizer. My doc didn’t even know what to do with the report. Took me 6 months to find a pharmacist who did. You gotta be your own advocate. Ask for PGx. Don’t wait for them to bring it up.
Aurora Memo
January 22, 2026 AT 22:37 PMThis is exactly why I push for pharmacogenomics in our clinic’s elderly care program. Not because it’s trendy. Because it saves lives. One patient stopped having falls after we switched her antidepressant based on her CYP2D6 status. She’s back gardening. That’s the kind of change we need more of.
chandra tan
January 23, 2026 AT 23:37 PMIn India, most doctors still think ‘genetics’ means ‘family history.’ They don’t know about CYP enzymes. But I’ve seen patients in rural clinics die from drug reactions because no one checked. We need community training. Not just labs. Real education. Maybe one day.
Dwayne Dickson
January 25, 2026 AT 15:29 PMLet’s be honest: if this were as simple as ‘just test everyone,’ we’d have done it by now. The real issue isn’t science-it’s bureaucracy. We’ve got AI models that can predict interactions with 37% more accuracy, but hospitals won’t integrate them because the EHR vendor charges extra. And don’t get me started on how ‘clinical decision support’ is often just a pop-up that says ‘see pharmacist’-with no pharmacist on staff. So yes, the tech works. The system? Still stuck in 1999.